Cloning and Sequencing of a Putative Sodium
Channel
Gene from a Parasitic Mite of the Honey
Bee, Varroa jacobsoni
Ruiwu Wang, Ke Dong and Zachary Huang
Department of Entomology, Michigan State University, East Lansing,
MI 48824, USA.
Introduction:
Voltage-gated sodium channels play a key role in mediating neural activity by allowing the rapid influx of sodium ions in the rising phase of action potentials. The major functional component of sodium channel is the a subunit, a large polypeptide (260KD) associated with two smaller subunits (b1 and b2) (Catterall, 1988). In insects, the a subunit is coded by the para locus, first identified in Drosophila (Loughney et al, 1989).
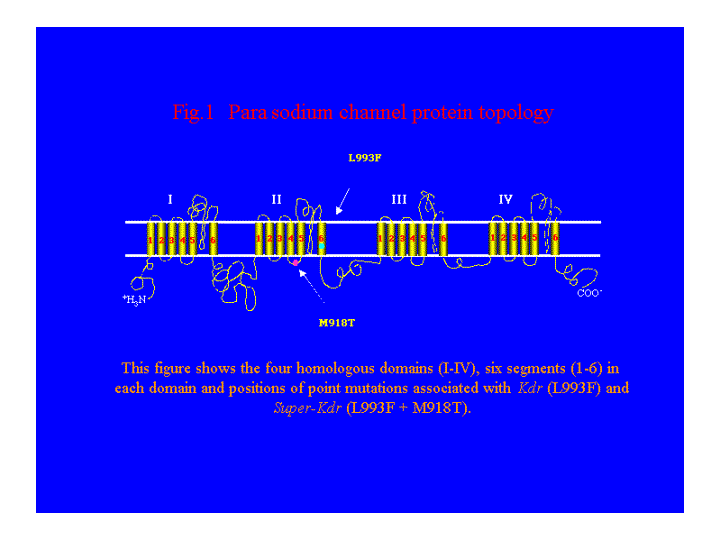
Voltage-gated sodium channels are the primary target sites of pyrethroid insecticides. A number of studies have shown that resistance to pyrethroid insecticides is associated with the para homologous sodium channel genes. In insect pests, such as house fly and cockroach, point mutations in the para homologous sodium channel gene are responsible for knockdown resistance (kdr), and super-kdr resistance to pyrethroids (Miyazaki et al, 1996; Dong, 1997; See Fig 1 ).

Varroa mite is the most serious pest of honey bees. Together with other pests and diseases, varroa mite caused colony mortality as high as 90-100% in recent years. Fluvalinate, a pyrethroid insecticide, has been used effectively to control the varroa mite. However, fluvalinate resistance has been detected in several states in US during the past two years. To study the resistance mechanisms of varroa mite to fluvalinate, we have partially cloned and sequenced mite cDNA of a para homologous sodium channel gene , using reverse transcriptase polymerase chain reaction (RT-PCR). We are determining whether mutations in para homologous sodium channel gene are associated with resistance to fluvalinate in the mite. It is hoped that by having a specific genetic marker, unique to mites that are resistant to fluvalinate, we will be able to determine whether or not mites are resistant to fluvalinate based on genetic screening of individual mites. This would be an alternative for the present field test (Pettis et al, 1998).
Materials and Methods:
Resistant mites (Maryland/R) were obtained from Maryland, which showed less than 80% mortality when treated with fluvalinate. No susceptible individuals were collected from Maryland. Mites from Michigan were separated into two groups: mites killed by fluvalinate during first 3 days were designated as susceptible (Michigan/S), those survived the treatment were collected with coumaphos and designated as resistant (Michigan/R).
Total RNA and poly (A) RNA were isolated from varroa mites, using RNA isolation kits from GIBCO/BRL according to the manufacturer’ instructions. First strand cDNA was synthesized using Superscript II RNase H- reverse transcriptase (GIBCO/BRL) in the presence of oligo(dT) or gene-specific primers. Degenerate primers were designed based on the conserved amino acid residues of sodium channels from Drosophila melanogaster, Blattella germanica, and Musca domestica, and were used for amplification of the homologous cDNA from the mites. PCR products were cloned and sequenced.
Results:
I.
Alignment of deduced amino acid sequence of there para sodium channel
protein in Varroa mites from Maryland and
Michighan
II. Table 1 Sequence similarity of the mite Para homologous sodium channel protein to other Para sodium channel proteins (IIS3-IVS6) (%)
|
microplus |
Germanica |
melanogaster |
domestica |
type II |
|
|
jacobsoni |
|
|
|
|
|
Conclusion:
1.
A partial para homologous sodium channel cDNA from fluvalinate-susceptible
and resistant mites was cloned and sequenced.
2. The deduced
amino acid sequence of domains II-IV of the mite para homologous sodium channel
shares 66.5%, 49.7%,
and 44.4% identity with the cattle tick (Boophilus microplus) para-homolog,
Drosophila para, and rat brain type II sodium
channel a-subunit respectively(See table 1).
3. There are
amino acid differences among mites from different locations. Howere, none
of these differences are associated with
mite resistance to pyrethroids. The kdr and super-kdr mutations were not
detected in mite homologous sodium channel gene
(See Fig.3).
4. Cloning and
sequencing of the full-length sodium channel cDNA sequence from varroa mite
are currently underway.
Reference:
1. Catterall WA (1988) Structure and
function of voltage-sensitive ion channel. Science 242: 50-61.
2. Loughney K, Kreber R, Ganetzky
B (1989) Molecular analysis of the para locus, sodium channel gene in Drosophila.
Cell 58: 1143-1154.
3. Dong K (1997) A single amino acid
changes in the para sodium channel protein associated with knockdown-resistance
(kdr)
to pyrethroid
insecticides in German cockroach. Insect Biochem Mol Biol 27: 647-654.
4. Miyazaki M, Ohyama K, Dunlap DY, Matsumura
F (1996) Cloning and sequencing of the para-type sodium channel gene
from susceptible
and kdr-resistant German cockroaches(Blattella germanica) and the house
fly (Musca domestica).
Mol Gen Genet 252:
61-68.
5. Pettis JS, Shimanuki H & Feldlaufer
MF (1998) Detecting fluvalinate-resistant varroa mites. American Bee J
138: 535-537.
We thank Jeff Pettis for providing the
mites from Maryland, Patti Elzen for mites from Florida (still being tested),
and Fred Dyer for mites from Michigan. This work was funded by a GREEEN
grant from the Michigan State University and a grant from the Plant and Pest
Management Division of Michigan Department of Agriculture.